|
Data Structures |
| struct | _data |
| | The _data struct holds a data set to be fit. More...
|
| struct | _meth |
| | The _meth struct determines which fitting method to call. More...
|
| struct | _func |
| | The _func struct holds pointers to the user-supplied functions. More...
|
| struct | _para |
| | The _para struct holds all of the paramters. More...
|
| struct | _fitf |
| | The gsl_fit struct will be passed as void pointer to the fitting routines. More...
|
Typedefs |
| typedef _fitf | gsl_fit |
| | The gsl_fit struct will be passed as void pointer to the fitting routines.
|
Enumerations |
| enum | _method { MULTIDIM_MINIMIZAION = 0,
LEAST_SQUARES = 1,
SIMULATED_ANNEAL = 2
} |
| | Flags for selecting the fiting method. More...
|
| enum | _algorithm {
CONJUGATE_FR = 0,
CONJUGATE_PR = 1,
VECTOR_BFGS = 2,
STEEPEST_DESCENT = 3,
LMSDR = 4,
LMDR = 5
} |
| | Flags for selecting the fiting algotrithm. More...
|
| enum | _type |
| | Flags for selecting whether single or multuple data sets are being fit. More...
|
Functions |
| gsl_fit * | gsl_fit_alloc (unsigned int method, unsigned int algotrithm, unsigned int max_iter, unsigned int type) |
| | Call this function to initate and zero the model.
|
| int | gsl_fit_add_dataset (gsl_fit *gft, double *xpt, double *ypt, double *sig, unsigned int num) |
| | Call this function to add a data set to be fit.
|
| int | gsl_fit_add_model_single (gsl_fit *gft, double(*fun)(double, double *), double *par, unsigned int *fix, unsigned int num) |
| | Call this function to add a model function when fitting single datasets.
|
| int | gsl_fit_add_model_multiple (gsl_fit *gft, double(*fun)(double, double *, unsigned int, unsigned int), double *par, unsigned int *fix, unsigned int num) |
| | Call this function to add a model function when fitting multiple datasets.
|
| int | gsl_fit_print (gsl_fit *gft, char *filename) |
| | Print out the current parameter fit.
|
| int | gsl_fit_do (gsl_fit *gft) |
| | Do the fit.
|
| int | gsl_fit_get_result (gsl_fit *gft, unsigned int function, double *par, double *err, unsigned int num) |
| | Get the results.
|
| void | gsl_fit_free (gsl_fit *gft) |
| | Free the gsl_fit.
|
| int | lstsq_main (void *void_gsl_fit_struct) |
| | The main function for performing method of least squares fitting.
|
| int | lstsq_fdf (const gsl_vector *parameters, void *void_gsl_fit_struct, gsl_vector *r, gsl_matrix *J) |
| | A routine which calculates the residuals 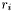 and the Jacobian matrix. and the Jacobian matrix.
|
| int | lstsq_f (const gsl_vector *parameters, void *void_gsl_fit_struct, gsl_vector *r) |
| | A routine which calculates the residuals 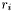 . .
|
| int | lstsq_df (const gsl_vector *parameters, void *void_gsl_fit_struct, gsl_matrix *J) |
| | A routine which calculates the Jacobian matrix.
|
| double | lstsq_derivatives (double p, void *void_gsl_fit_struct) |
| | A routine which calculates 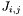 with a new parameter. with a new parameter.
|
| int | lstsq_status (gsl_fit *gft, int iteration, gsl_multifit_fdfsolver *s, gsl_matrix *c) |
| | A fitting status update printed to stderr.
|
| int | mdmin_main (void *void_gsl_fit_struct) |
| | The main function for performing multidimensional minimization.
|
| void | mdmin_fdf (const gsl_vector *parameters, void *void_gsl_fit_struct, double *f, gsl_vector *df) |
| | A routine which calculates the  value and the gradient of value and the gradient of  together. together.
|
| double | mdmin_f (const gsl_vector *parameters, void *void_gsl_fit_struct) |
| | A routine which calculates the  . .
|
| void | mdmin_df (const gsl_vector *parameters, void *void_gsl_fit_struct, gsl_vector *df) |
| | A routine which calculates the gradient of  . .
|
| double | mdmin_derivatives (double p, void *void_gsl_fit_struct) |
| | A routine which calculates  with a new parameter. with a new parameter.
|
| double | mdmin_chisq (gsl_fit *gft) |
| | A routine which calculates  . .
|
| void | mdmin_status (gsl_fit *gft, int iteration, gsl_multimin_fdfminimizer *s) |
| | A fitting status update printed to stderr.
|
| int | siman_main (void *void_gsl_fit_struct) |
| | The main function for performing simulated annealing.
|
| double | siman_energy (void *void_gsl_fit_struct) |
| | A routine which calculates  . .
|
| void | siman_step (const gsl_rng *rng, void *void_gsl_fit_struct, double step_size) |
| | Step the paramters by a small, random amount.
|
| void | siman_copy (void *source, void *dest) |
| | Copies one gsl_fit into another.
|
| void * | siman_construct (void *void_gsl_fit_struct) |
| | Creates a clone of a gsl_fit.
|
| void | siman_destroy (void *void_gsl_fit_struct) |
| | Frees a gsl_fit.
|
| double | siman_metric (void *void_gsl_fit_struct1, void *void_gsl_fit_struct2) |
| void | siman_print (void *void_gsl_fit_struct) |
| | Print out the current state of the solution.
|
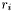 and the Jacobian matrix.
and the Jacobian matrix. 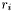 .
. 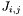 with a new parameter.
with a new parameter.  value and the gradient of
value and the gradient of  together.
together.  .
.  .
.  with a new parameter.
with a new parameter.  .
.  .
.  1.4.7
1.4.7