Functions
 .
. gsl_fit into another. gsl_fit. gsl_fit.
When using this technique, it can be useful to direct the stdout output to be saved in a file. It will contain the system energy and parameter values over the course of the simulation, which can be analyzed later.
Functions | |
| int | siman_main (void *void_gsl_fit_struct) |
| The main function for performing simulated annealing. | |
| double | siman_energy (void *void_gsl_fit_struct) |
A routine which calculates  . . | |
| void | siman_step (const gsl_rng *rng, void *void_gsl_fit_struct, double step_size) |
| Step the paramters by a small, random amount. | |
| void | siman_copy (void *source, void *dest) |
Copies one gsl_fit into another. | |
| void * | siman_construct (void *void_gsl_fit_struct) |
Creates a clone of a gsl_fit. | |
| void | siman_destroy (void *void_gsl_fit_struct) |
Frees a gsl_fit. | |
| double | siman_metric (void *void_gsl_fit_struct1, void *void_gsl_fit_struct2) |
| void | siman_print (void *void_gsl_fit_struct) |
| Print out the current state of the solution. | |
| int siman_main | ( | void * | void_gsl_fit_struct | ) |
| void_gsl_fit_struct | A void pointer, expected to be a gsl_fit. |
 , with respect to a set of parameters
, with respect to a set of parameters ![$\mathbf{a}=[a_1,a_2,\ldots]$](form_1.png) . The function
. The function  is given by,
is given by,
![\[ \chi^2 = \frac{1}{n-p} \sum_{i} \left(\frac{f(x_i;\mathbf{a}) - y_i}{\sigma_i}\right)^2 \]](form_35.png)
There are several parameters here that really should be configurable, but currently are not, such as the number of trial steps, and most importantly, the step size.
The initial temperature is set to be two times initial energy, which is just  evaluated with the initial parameters. The final temperature is
evaluated with the initial parameters. The final temperature is 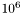 times smaller. This seems to work well when the Boltzmann constant is set to 1.
times smaller. This seems to work well when the Boltzmann constant is set to 1.
As in the other fitting methods, the system configuration is set to be the gsl_fit struct, so that the data is totally accessible. The parameters are overwritten with this technique, however, since the gsl_fit struct is used as workspace.
The difficulty is choosing the best step size, since there is only one that must be shared among all the variables.
Definition at line 12 of file gsl_siman.c.
00013 { 00014 double st, ft; 00015 gsl_rng *rng; 00016 gsl_siman_params_t params; 00017 gsl_fit *gft; 00018 00019 gft = (gsl_fit *)void_gsl_fit_struct; 00020 00021 rng = gsl_rng_alloc(gsl_rng_taus2); 00022 gsl_rng_set(rng, time(NULL)); 00023 00024 /* starting and final temperatures */ 00025 st = 2.0 * siman_energy(void_gsl_fit_struct); 00026 ft = st / 1.0e6; 00027 00028 /* 00029 ** The number of steps in temperature, n_T, is given by 00030 ** n_T = ln(st/ft)/ln(mu_t) 00031 ** 00032 ** The total number of trials, N, is given by 00033 ** N=n_T*iters_fixed_T 00034 */ 00035 00036 params.iters_fixed_T = 500; 00037 params.k = 1.0; 00038 params.t_initial = st; 00039 params.t_min = ft; 00040 params.mu_t = 1.01; 00041 params.step_size = 0.01; 00042 00043 fprintf(stderr, "Starting %d simulated annealing steps.\n\n", 00044 (int)floor(log(st/ft)/log(params.mu_t))); 00045 00046 gsl_siman_solve(rng, void_gsl_fit_struct, \ 00047 siman_energy, \ 00048 siman_step, \ 00049 siman_metric, \ 00050 siman_print, \ 00051 siman_copy, \ 00052 siman_construct, \ 00053 siman_destroy, \ 00054 0, params); 00055 00056 00057 gft->chis = siman_energy(void_gsl_fit_struct); 00058 00059 gsl_rng_free(rng); 00060 00061 return GSL_SUCCESS; 00062 }
| double siman_energy | ( | void * | void_gsl_fit_struct | ) |
| void_gsl_fit_struct | A void pointer, expected to be a gsl_fit. |
 .
. based on the parameters in
based on the parameters in gfc->para.par. Should be functionally identical with mdmin_chisq.
Definition at line 66 of file gsl_siman.c.
00067 { 00068 int i, j, k; 00069 double tmp; 00070 double residual = 0.0; 00071 int numdata; 00072 gsl_fit *gft; 00073 00074 gft = (gsl_fit *)void_gsl_fit_struct; 00075 00076 /* 00077 ** Loop over the data sets with index k. 00078 */ 00079 numdata = 0; 00080 for (k=0; k<gft->numd; k++) 00081 { 00082 /* 00083 ** Loop over the data points with index i 00084 */ 00085 for (i=0; i<gft->data[k].num; i++) 00086 { 00087 /* 00088 ** Loop over the functions with index j 00089 */ 00090 tmp = 0.0; 00091 for (j=0; j<gft->numf; j++) 00092 { 00093 if (gft->type == MULTIPLE) 00094 { 00095 tmp += gft->func[j].fun_m(gft->data[k].xpt[i], \ 00096 gft->para.par + gft->func[j].idx, \ 00097 k, j); 00098 } 00099 else 00100 { 00101 tmp += gft->func[j].fun_s(gft->data[k].xpt[i], \ 00102 gft->para.par + gft->func[j].idx); 00103 } 00104 } 00105 tmp = (tmp - gft->data[k].ypt[i]) / gft->data[k].sig[i]; 00106 residual += tmp * tmp; 00107 numdata++; 00108 } 00109 } 00110 tmp = residual / (numdata - gft->para.num); 00111 return tmp; 00112 }
| void siman_step | ( | const gsl_rng * | rng, | |
| void * | void_gsl_fit_struct, | |||
| double | step_size | |||
| ) |
| rng | A GSL random number generator | |
| void_gsl_fit_struct | A void pointer, expected to be a gsl_fit. | |
| step_size | The random stepsize. |
 .
.siman_main, and passed into this function as rng. The parameters are altered by a random value chosen in the interval ![$[-s,s]$](form_42.png) , where
, where 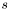 is the step size.
is the step size.
Definition at line 116 of file gsl_siman.c.
00117 { 00118 gsl_fit *gft; 00119 int i; 00120 double s; 00121 00122 gft = (gsl_fit *)void_gsl_fit_struct; 00123 00124 for (i=0; i<gft->para.num; i++) 00125 { 00126 if (gft->para.fix[i] == 0) 00127 { 00128 s = gsl_rng_uniform(rng); 00129 gft->para.par[i] = step_size * (2.0*s-1.0) + gft->para.par[i]; 00130 } 00131 } 00132 return; 00133 }
| void siman_copy | ( | void * | source, | |
| void * | dest | |||
| ) |
| source | A void pointer, expected to be a gsl_fit | |
| dest | A void pointer, expected to be a gsl_fit. |
gsl_fit into another.
Definition at line 137 of file gsl_siman.c.
00138 { 00139 int i, j; 00140 00141 gsl_fit *gft_old, *gft_new; 00142 00143 gft_old = (gsl_fit *)source; 00144 gft_new = (gsl_fit *)dest; 00145 00146 gft_new->numd = gft_old->numd; 00147 for (i=0; i<gft_old->numd; i++) 00148 { 00149 gft_new->data[i].idx = gft_old->data[i].idx; 00150 gft_new->data[i].num = gft_old->data[i].num; 00151 for (j=0; j<gft_old->data[i].num; j++) 00152 { 00153 gft_new->data[i].xpt[j] = gft_old->data[i].xpt[j]; 00154 gft_new->data[i].ypt[j] = gft_old->data[i].ypt[j]; 00155 gft_new->data[i].sig[j] = gft_old->data[i].sig[j]; 00156 } 00157 } 00158 00159 gft_new->para.num = gft_old->para.num; 00160 for (i=0; i<gft_old->para.num; i++) 00161 { 00162 gft_new->para.par[i] = gft_old->para.par[i]; 00163 gft_new->para.fix[i] = gft_old->para.fix[i]; 00164 gft_new->para.err[i] = gft_old->para.err[i]; 00165 gft_new->para.tmp[i] = gft_old->para.tmp[i]; 00166 } 00167 gft_new->para.idx = gft_old->para.idx; 00168 00169 gft_new->numf = gft_old->numf; 00170 for (i=0; i<gft_old->numf; i++) 00171 { 00172 gft_new->func[i].idx = gft_old->func[i].idx; 00173 gft_new->func[i].num = gft_old->func[i].num; 00174 gft_new->func[i].fun_s = gft_old->func[i].fun_s; 00175 gft_new->func[i].fun_m = gft_old->func[i].fun_m; 00176 } 00177 00178 gft_new->meth.solver_function_call = gft_old->meth.solver_function_call; 00179 gft_new->meth.mdmin_type = gft_old->meth.mdmin_type; 00180 gft_new->meth.lstsq_type = gft_old->meth.lstsq_type; 00181 00182 gft_new->chis = gft_old->chis; 00183 gft_new->maxi = gft_old->maxi; 00184 gft_new->indx = gft_old->indx; 00185 gft_new->type = gft_old->type; 00186 return; 00187 00188 }
| void* siman_construct | ( | void * | void_gsl_fit_struct | ) |
| void_gsl_fit_struct | A void pointer, expected to be a gsl_fit |
gsl_fit, cast as a void pointer.gsl_fit and clones the data from the old.
Definition at line 192 of file gsl_siman.c.
00193 { 00194 int i,j; 00195 00196 gsl_fit *gft_old, *gft_new; 00197 00198 gft_old = (gsl_fit *)void_gsl_fit_struct; 00199 gft_new = (gsl_fit *)malloc(sizeof(gsl_fit)); 00200 00201 gft_new->numd = gft_old->numd; 00202 gft_new->data = (struct _data *)malloc(gft_old->numd * sizeof(struct _data)); 00203 00204 for (i=0; i<gft_old->numd; i++) 00205 { 00206 gft_new->data[i].xpt = (double *)malloc(gft_old->data[i].num * \ 00207 sizeof(double)); 00208 gft_new->data[i].ypt = (double *)malloc(gft_old->data[i].num * \ 00209 sizeof(double)); 00210 gft_new->data[i].sig = (double *)malloc(gft_old->data[i].num * \ 00211 sizeof(double)); 00212 gft_new->data[i].idx = gft_old->data[i].idx; 00213 gft_new->data[i].num = gft_old->data[i].num; 00214 for (j=0; j<gft_old->data[i].num; j++) 00215 { 00216 gft_new->data[i].xpt[j] = gft_old->data[i].xpt[j]; 00217 gft_new->data[i].ypt[j] = gft_old->data[i].ypt[j]; 00218 gft_new->data[i].sig[j] = gft_old->data[i].sig[j]; 00219 } 00220 } 00221 00222 gft_new->para.num = gft_old->para.num; 00223 gft_new->para.par = (double *)malloc(gft_old->para.num*sizeof(double)); 00224 gft_new->para.fix = (unsigned int *)malloc(gft_old->para.num*\ 00225 sizeof(unsigned int)); 00226 gft_new->para.err = (double *)malloc(gft_old->para.num*sizeof(double)); 00227 gft_new->para.tmp = (double *)malloc(gft_old->para.num*sizeof(double)); 00228 for (i=0; i<gft_old->para.num; i++) 00229 { 00230 gft_new->para.par[i] = gft_old->para.par[i]; 00231 gft_new->para.fix[i] = gft_old->para.fix[i]; 00232 gft_new->para.err[i] = gft_old->para.err[i]; 00233 gft_new->para.tmp[i] = gft_old->para.tmp[i]; 00234 } 00235 gft_new->para.idx = gft_old->para.idx; 00236 00237 gft_new->numf = gft_old->numf; 00238 gft_new->func = malloc(gft_old->numf*sizeof(struct _func)); 00239 for (i=0; i<gft_old->numf; i++) 00240 { 00241 gft_new->func[i].idx = gft_old->func[i].idx; 00242 gft_new->func[i].num = gft_old->func[i].num; 00243 gft_new->func[i].fun_s = gft_old->func[i].fun_s; 00244 gft_new->func[i].fun_m = gft_old->func[i].fun_m; 00245 } 00246 00247 gft_new->meth.solver_function_call = gft_old->meth.solver_function_call; 00248 gft_new->meth.mdmin_type = gft_old->meth.mdmin_type; 00249 gft_new->meth.lstsq_type = gft_old->meth.lstsq_type; 00250 00251 gft_new->chis = gft_old->chis; 00252 gft_new->maxi = gft_old->maxi; 00253 gft_new->indx = gft_old->indx; 00254 gft_new->type = gft_old->type; 00255 00256 return (void *)gft_new; 00257 }
| void siman_destroy | ( | void * | void_gsl_fit_struct | ) |
| void_gsl_fit_struct | A void pointer, expected to be a gsl_fit |
gsl_fit . SHould be functionally identical to gsl_fit_free
Definition at line 261 of file gsl_siman.c.
00262 { 00263 int i; 00264 gsl_fit *gft; 00265 00266 if (void_gsl_fit_struct == NULL) return; 00267 00268 gft = (gsl_fit *)void_gsl_fit_struct; 00269 00270 for(i=0; i<gft->numd; i++) 00271 { 00272 free(gft->data[i].xpt); 00273 free(gft->data[i].ypt); 00274 free(gft->data[i].sig); 00275 } 00276 if (gft->numd) free(gft->data); 00277 00278 if (gft->para.num) 00279 { 00280 free(gft->para.par); 00281 free(gft->para.fix); 00282 free(gft->para.err); 00283 free(gft->para.tmp); 00284 } 00285 00286 if (gft->numf) 00287 { 00288 free(gft->func); 00289 } 00290 00291 free(void_gsl_fit_struct); 00292 00293 return; 00294 }
| double siman_metric | ( | void * | void_gsl_fit_struct1, | |
| void * | void_gsl_fit_struct2 | |||
| ) |
| void_gsl_fit_struct1 | A void pointer, expected to be a gsl_fit | |
| void_gsl_fit_struct2 | A void pointer, expected to be a gsl_fit |
Definition at line 298 of file gsl_siman.c.
00299 { 00300 gsl_fit *gft1; 00301 gsl_fit *gft2; 00302 double sum, tmp; 00303 int i; 00304 00305 gft1 = (gsl_fit *)void_gsl_fit_struct1; 00306 gft2 = (gsl_fit *)void_gsl_fit_struct2; 00307 sum = 0.0; 00308 for (i=0; i<gft1->para.num; i++) 00309 { 00310 /*sum += fabs(gft1->para.par[i] - gft2->para.par[i]);*/ 00311 tmp = (gft1->para.par[i] - gft2->para.par[i]); 00312 sum += tmp * tmp; 00313 } 00314 return sqrt(sum); 00315 }
| void siman_print | ( | void * | void_gsl_fit_struct | ) |
| void_gsl_fit_struct | A void pointer, expected to be a gsl_fit |
stdout the current parameter values. The printf statement adds columns to the output of the gsl_siman_solve routine. The n columns of the output are: 1: Number of iterations, i.e. number of times the temperature has been reduced. 2: Number of evaluations, equal to the number of iterations times the number trials at each temperature 3: The temperature 4 through n-1: The parameters n: The energy
Definition at line 319 of file gsl_siman.c.
00320 { 00321 gsl_fit *gft; 00322 int i; 00323 00324 gft = (gsl_fit *)void_gsl_fit_struct; 00325 00326 for (i=0; i<gft->para.num; i++) 00327 { 00328 printf(" %10.5g ", gft->para.par[i]); 00329 } 00330 return; 00331 }
 1.4.7
1.4.7