| int gsl_fit_add_dataset | ( | gsl_fit * | gft, | |
| double * | xpt, | |||
| double * | ypt, | |||
| double * | sig, | |||
| unsigned int | num | |||
| ) |
- Parameters:
-
gft A pointer to a gsl_fitxpt Array of 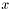 data
data ypt Array of 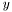 data
data sig Array of errors in the 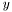 data
data num Length of data arrays
- Returns:
- GSL error code.
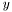 data, simply set
data, simply set 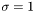 .
.
Definition at line 114 of file gsl_fit.c.
00117 { 00118 int i; 00119 00120 /* Sanity checks */ 00121 if (gft==NULL) { 00122 GSL_ERROR("Attempt to add data to null gsl_fit", \ 00123 GSL_EFAULT); } 00124 if ((gft->type==SINGLE)&&(gft->numd!=0)) { 00125 GSL_ERROR("Already have data to fit", \ 00126 GSL_EINVAL); } 00127 if (num<=2) { 00128 GSL_ERROR("Not enough data points", \ 00129 GSL_EINVAL); } 00130 00131 /*==============*/ 00132 /* allocate the memory for the */ 00133 gft->data = realloc(gft->data, (gft->numd+1)*sizeof(struct _data)); 00134 if (gft->data == NULL) { 00135 GSL_ERROR("No memory for data set",\ 00136 GSL_ENOMEM); } 00137 00138 /*==============*/ 00139 gft->data[gft->numd].xpt = (double *)malloc(num*sizeof(double)); 00140 gft->data[gft->numd].ypt = (double *)malloc(num*sizeof(double)); 00141 gft->data[gft->numd].sig = (double *)malloc(num*sizeof(double)); 00142 if ((gft->data[gft->numd].xpt == NULL) || \ 00143 (gft->data[gft->numd].ypt == NULL) || \ 00144 (gft->data[gft->numd].sig == NULL)) { 00145 GSL_ERROR("No memory for data set",\ 00146 GSL_ENOMEM); } 00147 00148 /*==============*/ 00149 gft->data[gft->numd].num = num; 00150 gft->data[gft->numd].idx = 0; 00151 00152 /*==============*/ 00153 for (i=0; i<num; i++) 00154 { 00155 gft->data[gft->numd].xpt[i] = xpt[i]; 00156 gft->data[gft->numd].ypt[i] = ypt[i]; 00157 gft->data[gft->numd].sig[i] = sig[i]; 00158 } 00159 00160 /*==============*/ 00161 gft->numd += 1; 00162 00163 /*==============*/ 00164 return GSL_SUCCESS; 00165 }
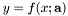 , i.e., for a single point
, i.e., for a single point 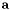 , return the value
, return the value  1.4.7
1.4.7